
The Benefits of At-Home Genetic Testing For Celiac Disease
At-home genetic testing for celiac disease is a groundbreaking innovation helping people to determine their

GlutenID® testing was featured in a recent article published by the College of American Pathologists (CAP) monthly newsletter called CAP Today.
CAP TODAY and the Association for Molecular Pathology have teamed up to bring molecular case reports to CAP TODAY readers. AMP members write the reports using clinical cases from their own practices that show molecular testing’s important role in diagnosis, prognosis, and treatment. The following report comes from PacificDx clinical laboratory, Irvine, Calif. If you would like to submit a case report, please send an email to the AMP at amp@amp.org. For more information about the AMP and all previously published case reports, visit www.amp.org.
Shelly Gunn, MD, PhD
Mathew W. Moore, PhD
Philip D. Cotter, PhD
Dermatitis herpetiformis (DH) is an autoimmune skin disorder associated with celiac disease and characterized by pruritic, blistering lesions mainly on the elbows, knees, buttocks, lower back, and scalp. Immunopathogenesis is attributed to IgA TG3 antibody immune complexes in the papillary dermis originating from subclinical celiac disease in the gut.1 Onset occurs at any age, but DH is diagnosed most often in young adults and rarely in children. Individuals of northern European heritage represent the most frequently affected population for DH, with an estimated incidence reported by the National Organization for Rare Disorders of 75.3 per 100,000 people.2 First-degree family members of patients with DH are at increased risk for both DH and celiac disease.3
The cause of DH and celiac disease is multifactorial and includes a strong genetic component, specifically the presence of genetic risk haplotypes HLA-DQ2 and/or HLA-DQ8 encod-ed by variants of the HLA-DQA1 and HLA-DQB1 genes on chromosome 6.4 Diagnosis of DH can be made in patients with dermatologic and/or GI-related symptoms using perilesional skin biopsy for direct immunofluores-cence microscopy to reveal pathognomonic IgA depositions in the papil-lary dermis.1 Diagnosis is supported by IgA anti-tissue transglutaminase (tTG) antibody serological testing and villous atrophy identified with endoscopic small bowel biopsy.5
Tissue biopsy and tTG antibody testing for specific IgA class circulating antibodies are accurate for diagnosing DH and celiac disease when the patient is consuming dietary gluten to trigger active disease. Traditional confirmatory testing has a high false-negative rate in patients who have eliminated gluten from their diet resulting in quiescent disease. For confirmation of DH and celiac disease in “gluten free” patients, some physicians have used for diagnostic purposes a “gluten challenge,” in which the patient adds gluten back into the diet for four to eight weeks to reactivate disease in preparation for biopsy and antibody testing.6 This approach is disruptive for asymptomatic individuals, however, and needlessly invasive if they do not carry the DQ2 or DQ8 genes. In the case we report here, we discuss the use of next-generation sequencing as an accurate, one-time, noninvasive, celiac genetic risk assessment screening for the family of a child in remission from DH following elimination of gluten from his diet.
Case. A previously healthy 21-month-old child presented with a chronic symmetrical blistering pruritic rash covering both buttocks and mild GI symptoms that appeared shortly after introduction of solid foods including wheat products (Fig. 1) The rash persisted for several weeks and was evaluated via pediatric telemedicine because of the COVID-19 lockdown. The presumptive diagnosis was allergic reaction to a cat (even though the family did not own one), and topical steroids were initiated without improvement. His grandmother, an anatomic and clinical pathologist, recognized the characteristic DH lesions and suggested trial of a gluten-free diet. She had a high index of suspicion because of her son’s (the child’s father) history of chronic GI symptoms, which had never been diagnosed as celiac disease. Within a few weeks of eliminating gluten the child was completely asymptomatic. Serologic tTG and endomysial anti-body testing performed after dietary gluten was eliminated was found to be negative for circulating IgA.
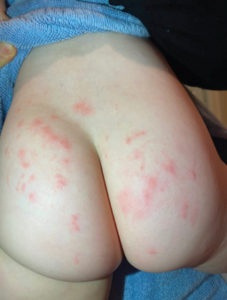
The family is of northern European origin and due to concern for the patient’s two asymptomatic siblings (ages four years and 10 months), family celiac genetic health risk studies were ordered.7 The testing was performed using PacificDx’s GlutenID® test, a next-generation sequencing assay designed to analyze variants associated with HLA-DQA1 and HLA-DQB1 genes for identification of haplotypes known to fall within a spectrum of risk for celiac disease. The test analyzes all possible celiac genetic risk haplotypes supported by level one (the highest) level of evidence. If none of the celiac risk-associated haplotypes are detected, the result has a negative predictive value for celiac disease of greater than 90 percent.
Materials and methods. The laboratory extracted DNA from buccal cell swabs collected at home by all five family members and analyzed it using Illumina MiSeqDx technology. HLA-tagging celiac risk single nucleotide polymorphisms for DQ2.5, DQ8, DQ2.2, and DQ7 were amplified and sequenced at a minimum of 40× coverage. The test identified heterozygous and homozygous combinations of the celiac risk alleles for each family member, and these were reported within a spectrum of risk from low to elevated. When none of the celiac risk alleles were identified, the results were reported as nonceliac genetics (NCG).
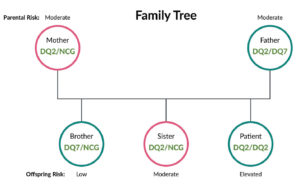
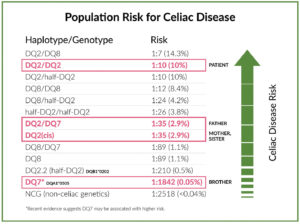
Results. The homozygous DQ2/ DQ2 GlutenID® associated with elevated celiac disease risk was identified in the patient. Each of his parents was found to carry one DQ2 and his father also had a DQ7 allele, placing both parents at moderate risk. His four year old sister inherited the paternal DQ2 and maternal NCG, placing her at moderate risk also. His younger brother inherited DQ7 from the paternal chromosome and a maternal NCG, placing him on the lowest end of the risk spectrum7 (Figs. 2 and 3).
Discussion. Approximately 30 to 40 percent of the world population carries specific allelic combinations of DQ2/DQ8, placing them on a spectrum of risk for celiac disease. It is well supported in the medical litera-ture that the presence of DQ2/DQ8 genes is necessary but not sufficient for development of celiac disease, and the negative predictive value of negative DQ2/DQ8 test results is greater than 90 percent. Thus, an individual who tests negative for celiac risk alleles is highly unlikely to have celiac disease or benefit from invasive diagnostic procedures, except in the rare scenario of high clinical suspicion of celiac disease in the presence of negative genetics.
In a recent large-scale pediatric screening study of 9,973 children ages one to 17 years, a high prevalence of undiagnosed celiac disease was confirmed in both asymptomatic and symptomatic participants.11 A limitation of the study was the use of IgA tTG antibody testing as the screening method, which can produce false-negative results in high-risk children who have not consumed adequate gluten before the test. Using a one-time, DNA-based, celiac genetic health risk test may become a preferred screening method in pediatric populations for identifying those who are genetically at risk and would benefit from repeated antibody screening as well as those without risk alleles who can forgo such testing. As a screening method, NGS has advantages over other celiac disease–associ-ated HLA typing methods, such as allele-specific PCR and Sanger sequencing. These include the cost-effectiveness of multiplexing samples for family screening and scalability for large-population applications, such as mass screening in high-risk groups.12
In this case, the patient was suspected to have subclinical celiac disease based on his dermatologic findings and had complete reversal of symptoms on a gluten-free diet. Retrospective molecular testing identified the DQ2/DQ2 homozygous genotype correlated with strongly increased risk for celiac disease development due to a phenotype of enhanced gluten antigen presentation to the immune system. 13,14 The child’s genotype likely contributed to the rare presentation of DH at such a young age. As the patient was in remission after removal of dietary gluten, the family elected to forgo gluten challenge and invasive confirmatory testing. However, launching a toddler on a lifetime glu-tenfree diet is best approached with objective evaluation of the clinical evidence in consultation with the family’s health care providers. If the child’s symptoms recur due to dietary noncompliance or other reasons, there would be an opportunity to repeat the antibody testing and proceed with tissue biopsies. The hope for this child and his family, however, is that knowledge of their celiac genetic risk will result in a lifetime of informed dietary choices based on a shared commitment to wellness.
1. Reunala T, Salmi TT, Hervonen K. Dermatitis herpetiformis: pathognomonic transglutaminase IgA deposits in the skin and excellent prognosis on a gluten-free diet. Acta Derm Venereol. 2015; 95(8):917–922. 2. Dermatitis herpetiformis. National Organiza-tion for Rare Disorders. https://rarediseases.org/ rare-diseases/dermatitis-herpetiformis. 3. Nellikkal SS, Hafed Y, Larson JJ, Murray JA, Absah I. High prevalence of celiac disease among screened first-degree relatives. Mayo Clin Proc. 2019; 94(9):1807–1813. 4. Megiorni F, Pizzuti A. HLA-DQA1 and HLA-DQB1 in celiac disease predisposition: practical implications of the HLA molecular typing. J Biomed Sci. 2012;19(1):88. 5. Lau MS, Sanders DS. Optimizing the diagnosis of celiac disease. Curr Opin Gastroenterol. 2017; 33(1):173–180. 6. Leonard J, Haffenden G, Tucker W, et al. Gluten challenge in dermatitis herpetiformis. N Engl J Med. 1983;308(14):816–819. 7. Singh P, Arora S, Lal S, Strand TA, Makharia GK. Risk of celiac disease in the first- and second-degree relatives of patients with celiac disease: a systematic review and meta-analysis. Am J Gastroenterol. 2015; 110(11):1539–1548. 8. Pietzak MM, Schofield TC, McGinnis MJ, Na-kamura RM. Stratifying risk for celiac disease in a large at-risk United States population by using HLA alleles. Clin Gastroenterol Hepatol. 2009;7(9):966–971. 9. Megiorni F, Mora B, Bonamico M, et al. HLA-DQ and risk gradient for celiac disease. Hum Immunol. 2009;70(1):55–59. 10. Taylor AK, Lebwohl B, Snyder CL, Green P. Celiac disease. GeneReviews; 2008. Updated Jan. 31, 2019. https://www.ncbi.nlm.nih.gov/books/ NBK1727. 11. Stahl MG, Rasmussen CG, Dong F, et al. Mass screening for celiac disease: the autoimmunity screening for kids study. Am J Gastroenterol. 2021; 116(1):180–187. 12. Ludvigsson JF, Card TR, Kaukinen K, et al. Screening for celiac disease in the general population and in high-risk groups. United European Gastroen-terol J. 2015;3(2):106–120. 13. Vader W, Stepniak D, Kooy Y, et al. The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses. Proc Natl Acad Sci USA. 2003;100(21):12390–12395. 14. De Silvestri A, Capittini C, Poddighe D, et al. HLA-DQ genetics in children with celiac disease: a meta-analysis suggesting a two-step genetic screen-ing procedure starting with HLA-DQ β chains. Pediatr Res. 2018;83(3):564–572.
Dr. Gunn is chief medical officer of Re-searchDx and medical director of Paci-ficDx clinical laboratory, a subsidiary of ResearchDx, and Dr. Moore and Dr. Cotter are principals and co-founders of ResearchDx—all in Irvine, Calif.

At-home genetic testing for celiac disease is a groundbreaking innovation helping people to determine their

Virtually everyone prefers a simple cheek swab to an invasive blood test but that’s not

GlutenID® testing was featured in a recent article published by the College of American Pathologists